Our laboratory aims to identify and further investigate the role of mechanical forces in cellular behavior and signaling, with the ultimate goal to improve human health and health care.
Our research has a central focus on
cellular mechanobiology signaling in particular cell-matrix interactions that guide the control of gene expression and regulation. From our work there are two main clinically applied areas:
- Applied Immuno-Mechanobiology: Mechanobiological signaling associated with lymphocyte activation and expansion.
- Applied Neuro-Mechanobiology: Protein aggregation and phase transition in motor neuronal disease. The aim is to find molecular ways for intervention, and the development of next generation therapies.
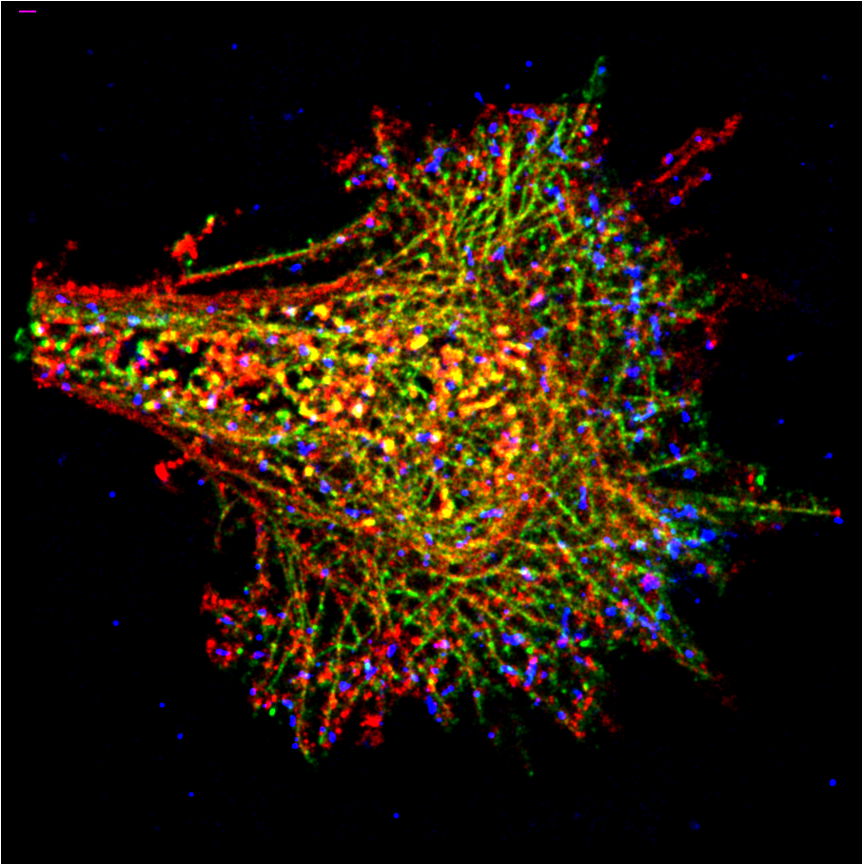
In our interdisciplinary laboratory, besides cell culture we have a strong imaging core, which allows the development and application of multi-scale imaging techniques to quantify and characterize tissues, cells, single proteins, extracellular matrix, and cell-matrix interactions answer our mechanobiology research questions.
Our laboratory core techniques:
Biophysics: Single Molecule Localization Microscopy (STORM/PALM), high throughput SMLM, Structured Illumination Microscopy, STED, Lattice Light Sheet Microscopy, Airyscan, 2-Photon Microscopy, Traction Force Microscopy, single molecule FRET force sensors, MEMS devices, micro and nanostructuring, TOCCSL, FRAP, AFM
Biochemistry: surface coating, gene engineering, transduction, transfection, protein conjungation, ELISA, protein purification, gene expression, single cell transcriptomics, cell lines, immunological assays, GMP conform production, phenotyping and activation analysis, flow cytometry, FACS, DNA FISH.
 Together with my research groups in Berlin and Zurich we investigate mechanobiological pathways and external cues that influence T cell and B cell signaling to tune their consequent functions. Here we explore how different conditions such as surface stiffness, topography, and hydrostatic pressure influence lymphocytes.
Together with my research groups in Berlin and Zurich we investigate mechanobiological pathways and external cues that influence T cell and B cell signaling to tune their consequent functions. Here we explore how different conditions such as surface stiffness, topography, and hydrostatic pressure influence lymphocytes.
 Our laboratory is associated with a joint international research initiative, which aims to better understand protein aggregation and phase transition toward motor neuronal disease prevention, intervention, and the development of next generation therapies.
Our laboratory is associated with a joint international research initiative, which aims to better understand protein aggregation and phase transition toward motor neuronal disease prevention, intervention, and the development of next generation therapies.